Research
Gene-isoform diversity and post-transcriptional regulation
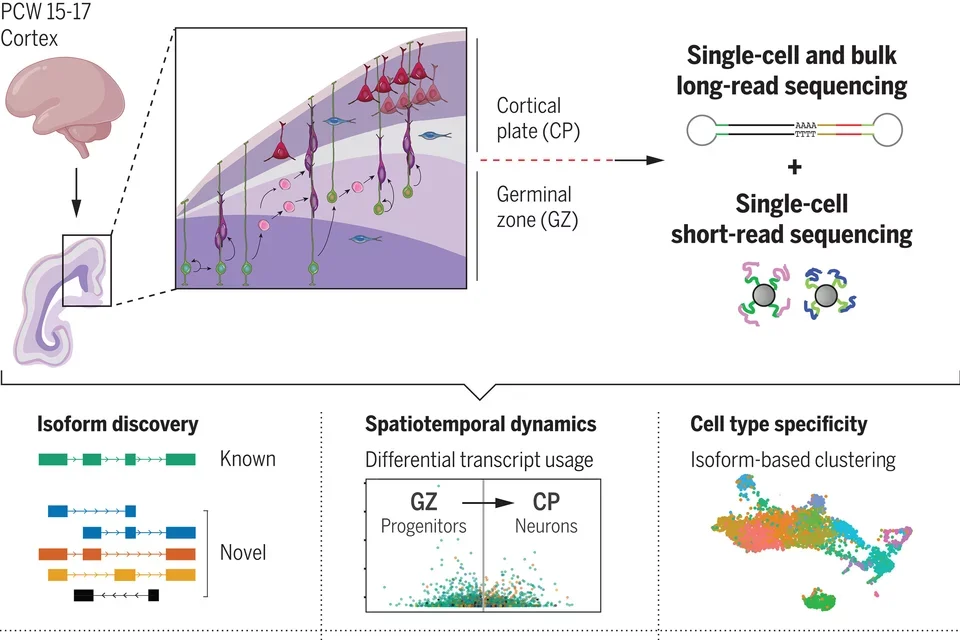
The development of the human brain is regulated by precise molecular mechanisms driving spatiotemporal and cell type–specific transcript expression programs. RNA splicing and RNA editing, as well as other post-transcriptional regulatory mechanisms are highly prevalent in the brain, yet their role in brain development and function is not well understood. In a recent study (Science, 2024) we leveraged single-molecule long-read sequencing to deeply profile the full-length transcriptome of the germinal zone and cortical plate regions of the developing human neocortex at tissue and single-cell resolution. We uncovered thousands of novel gene-isoforms with distinct spatiotemporal expression patterns predicted to regulate neurogenesis and highlighted the importance of this form of regulation in neurodevelopmental disorders. In collaboration with the Stein laboratory, we have also investigated how genetic variation impacts RNA editing (AJHG, 2024) and miRNA regulation (Elife, 2023) during neurogenesis. These foundational studies highlight the importance of understanding the mechanisms regulating isoform diversity in the brain.
Mechanisms of corticogenesis, cortical expansion and evolution
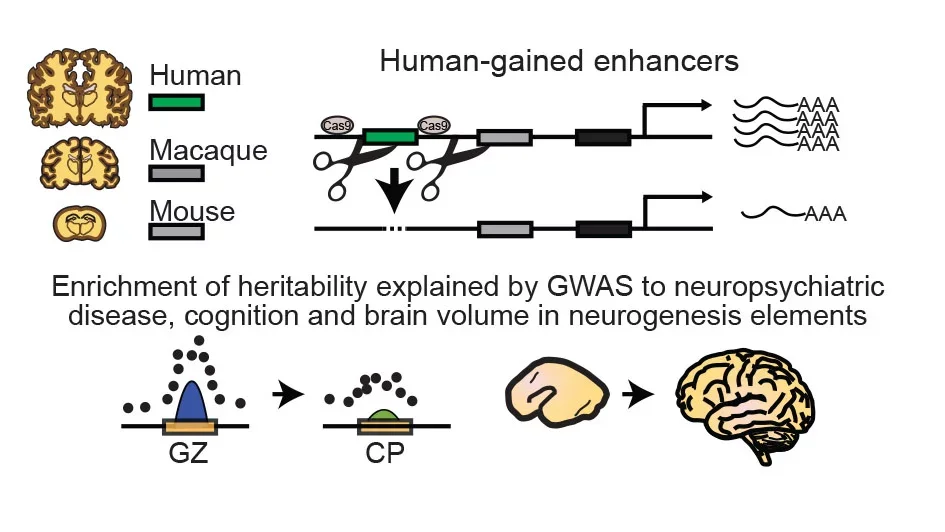
What makes us human? The neocortex is responsible for many higher-order, uniquely human cognitive and social capabilities. Comparative cross-species studies indicate that the expansion of the neocortex is a central mechanism in the evolution of human cognition. Recent efforts have identified human-specific genetic variation and present an invaluable opportunity to identify molecular mechanisms guiding neocortical expansion and other human-specific neurodevelopmental processes. Understanding this process is hindered by incomplete knowledge of the cells, gene expression patterns, and gene regulatory mechanisms acting during human cortical neurogenesis. Leveraging Hi-C, ATAC-seq, single-cell genomics, and long-read sequencing, we study the cell-specific genetic programs of the developing human neocortex (Nature, 2016; Cell, 2018; Neuron, 2019; Science 2024). We have discovered (Cell, 2018) human-gained enhancers that regulate genes enriched in outer radial glia (oRG), a cell type is particularly expanded in the developing cortex of highly gyrencephalic species, including humans. Our data suggests that human-specific variation drives oRG development and is a critical element in human brain evolution. We also identified FGFR2 as target of a human-gained enhancer and showed using CRISPR/Cas9 genomic engineering, that deletion of this enhancer disrupts cortical neurogenesis. We are actively studying the mechanisms driving human cortical evolution.
Mapping gene regulation in developing human neocortex
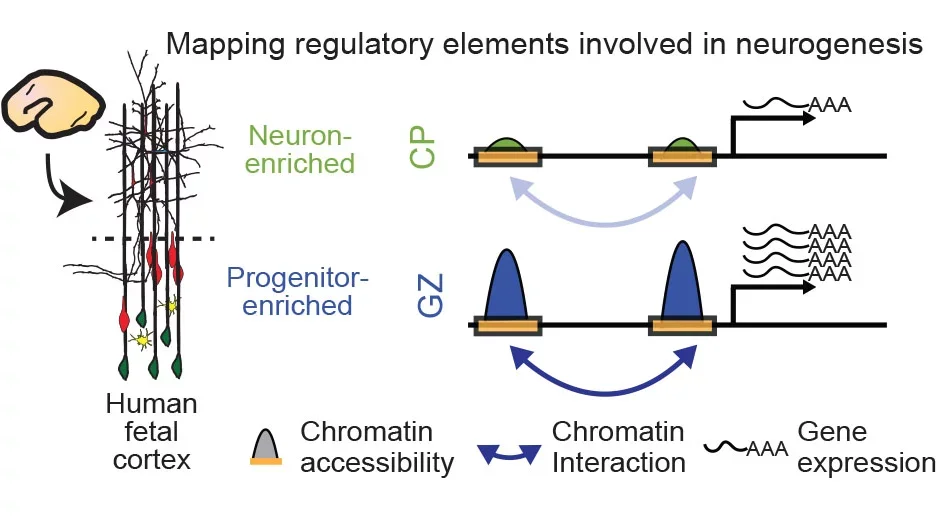
How does the genome direct the production of the 32 billion cells in the human cortex? The generation of neurons and subtypes of glia in the human neocortex is achieved through tightly regulated control of proliferation and differentiation of neural progenitors. We are interested in the cellular and molecular mechanisms orchestrating cortical neurogenesis, in particular in how non-coding regulatory elements interact with transcription factors and chromatin modifiers to regulate gene expression and direct neural progenitor proliferation and differentiation. We analyzed the dynamic changes in chromatin structure to identify non-coding regulatory elements and transcription factors that direct key genes involved in neurogenesis (Nature, 2016; Cell, 2018). We used this data to create a comprehensive map of regulatory elements and their target genes. In collaboration with other groups, we have extended this work to study how genetic variation regulates neurogenesis in vivo (Cell, 2019) and with cell type specificity (Nat. Neuro., 2021). With these resources, we can now assess how genetic variation in these non-coding regions acts on specific pathways, cell types and biological processes during cortical development.
Mechanisms of neuropsychiatric disorders
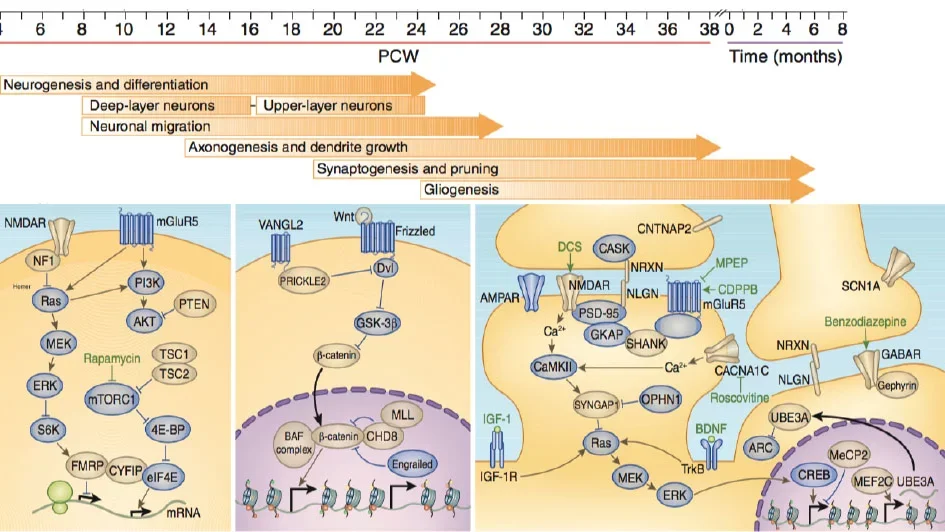
Multiple lines of evidence suggest that the developing human neocortex is particularly susceptible to genetic risk for neuropsychiatric disease, including autism spectrum disorder (ASD) and schizophrenia (see our Nat. Med. Review). We have found that de novo variants causing high risk for ASD are enriched in genes co-expressed during early neurogenesis in human cortex in developing neurons (Neuron, 2019) and that those processes are recapitulated in our human neural stem models (Neuron, 2014). More recently, we have found that high risk ASD genes and those implicated in neurodevelopmental disorders are more likely to encode multiple isoforms with specific cell-type expression (Science, 2024) in the developing neocortex. Genetic risk for schizophrenia appears to act in processes and cell types enriched in the germinal zone (GZ) of the human neocortex, which harbors neural progenitors and early-born neurons. Notably, genetic variation associated with human cognitive performance is also preferentially enriched in the GZ (Cell, 2018). We are interested in using multiple approaches including gene expression, chromatin accessibility profiling and genomic engineering in human neural stem cells to identify how corticogenesis and normal brain function are dysregulated in neuropsychiatric disorders.
Modeling human brain development and function using stem cells
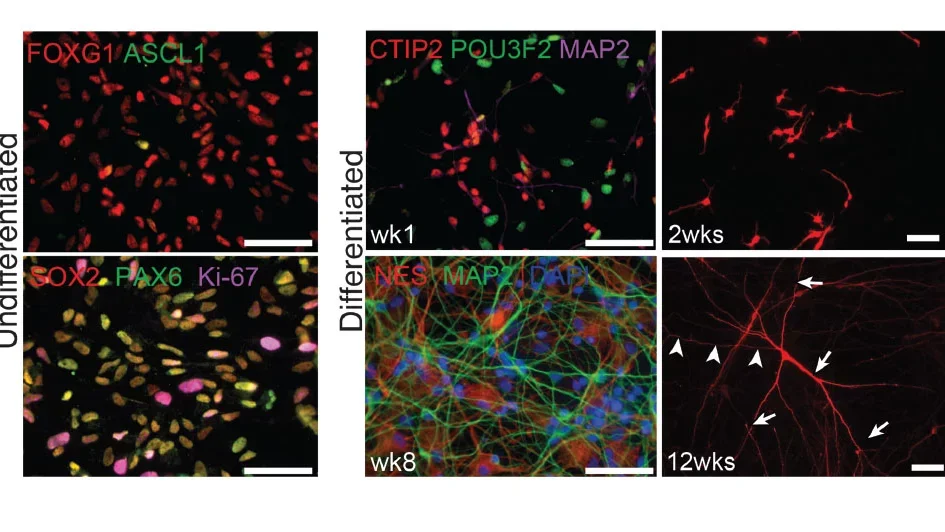
Advances in neural stem cell biology are providing for the first time an ideal platform for mechanistic studies in human tissue that bypasses species-specific differences in gene expression and cellular mechanisms. A major gap in the study of human neural stem cell models is a lack of quantitative approaches and thorough assessment of their fidelity to in vivo brain development. We developed CoNTExT, an unbiased quantitative approach to assess the capacity of human in vitro systems to model brain development using transcriptional profiling. CoNTExT enables us to determine the neuroanatomical identity and developmental maturity of neural stem cell cultures, as well as allowing us to identify signaling mechanisms preserved in vitro. CoNTExT is a valuable tool for the scientific community to assess the fidelity of in vitro cultures, including tri-dimensional and organoid cultures, and revealed key differences and limitations of different models. Our work on CoNTExT was featured on the cover of Neuron. A major interest of our lab is to continue developing, improving and characterizing neural stem cell models, harnessing their potential to elucidate mechanisms of normal brain development and function.